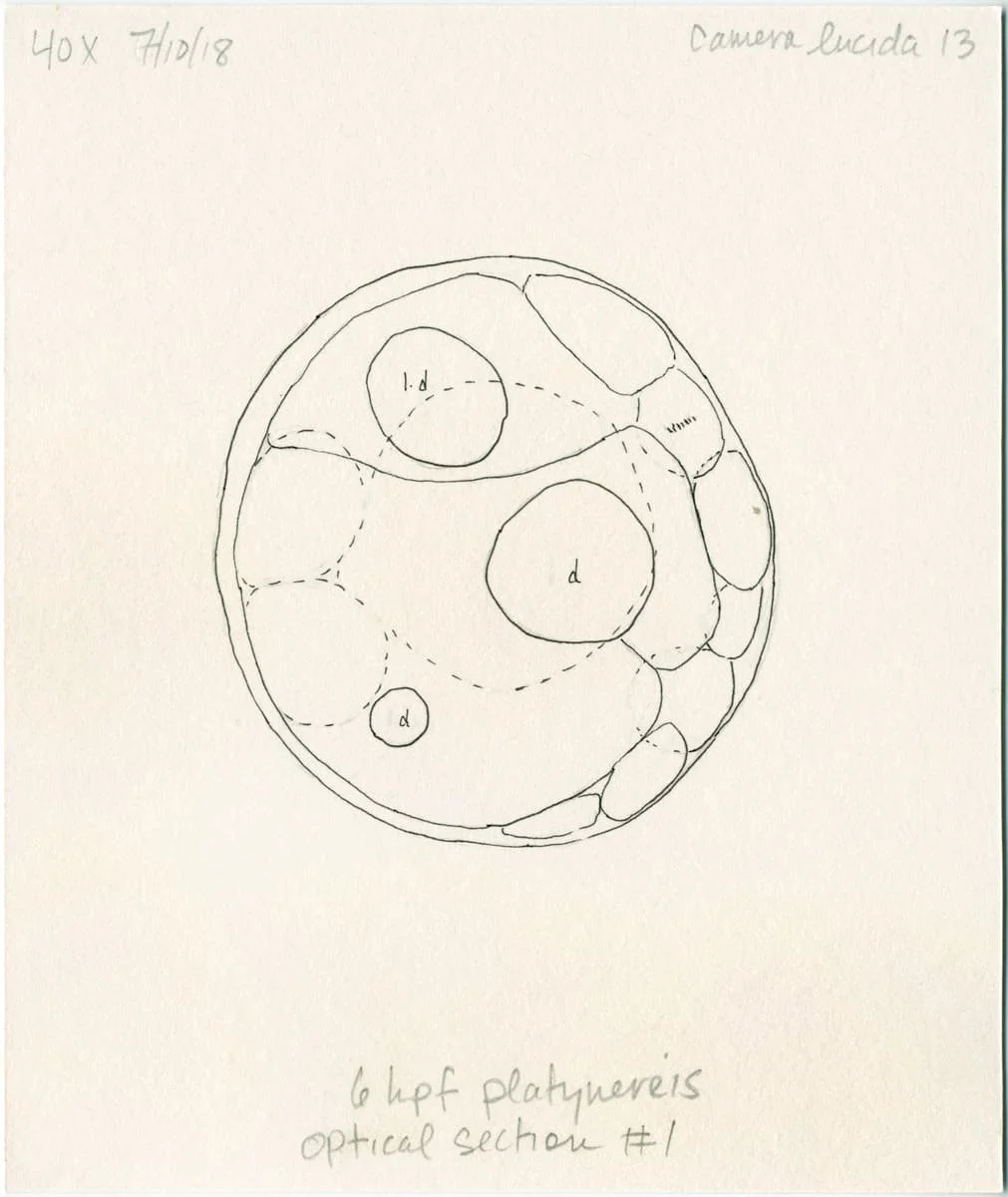
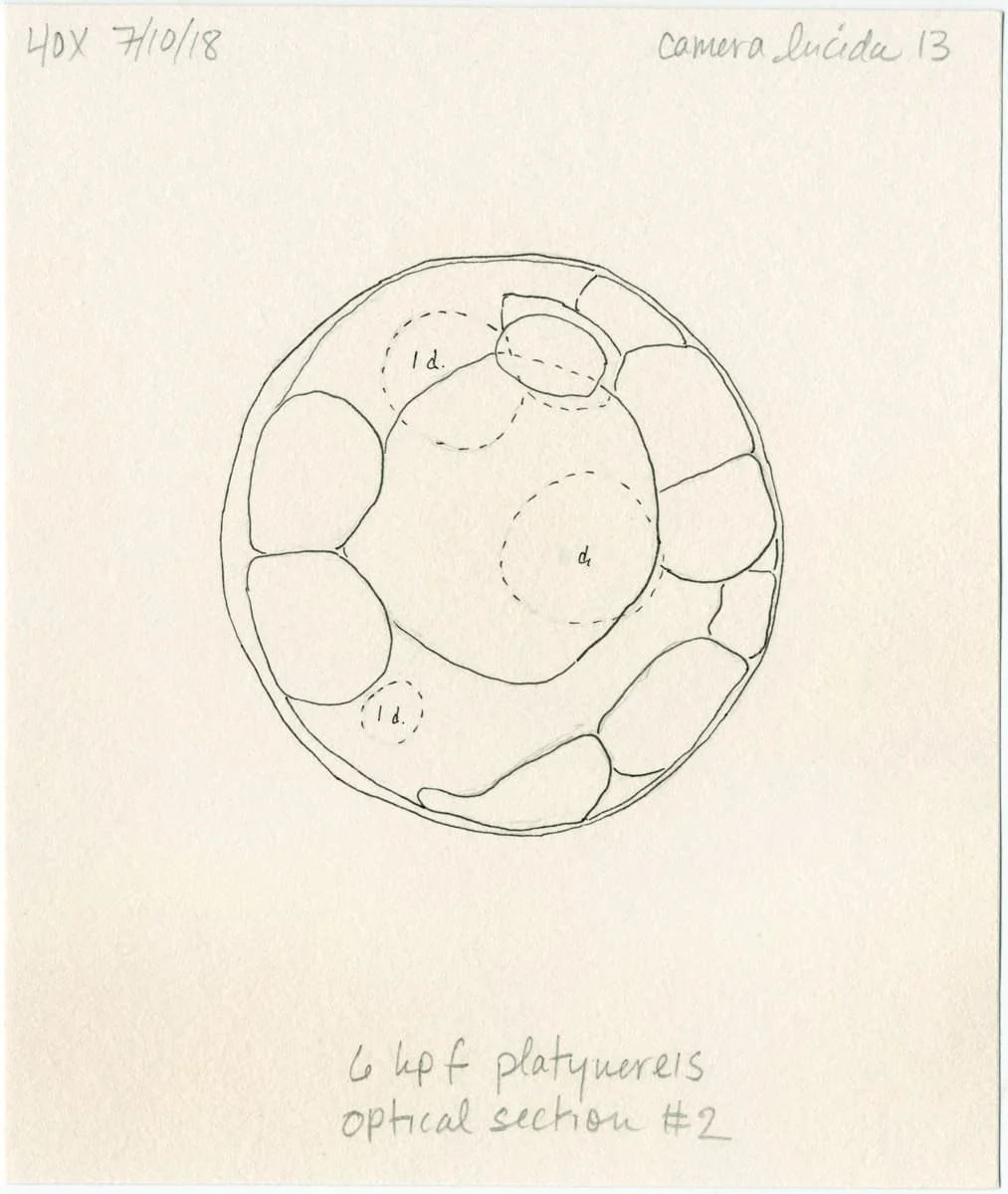
Morphogenesis
A 3D rendered 32-cell Platynereis dumerilii embryo.
Most of the organisms we encounter on a daily basis emerged from a single cell — an egg, seed, or spore — or small cluster of cells. My research explores morphogenesis, the developmental process through which the forms of multicellular organisms emerge. I look at the dynamic mechanisms of morphogenesis that arise from interactions among molecules, genes, cells, and structurally integrated tissues. I’m particularly interested in how the geometries and mechanics of cells carry information that regulates cell-fate determination as well as how the cytoskeleton (actin, non-muscle myosin, and microtubules) physically sculpts the shapes of organisms. I primarily work with marine invertebrates due to the relative transparency and beauty of their embryos, and the range of developmental strategies they have evolved.
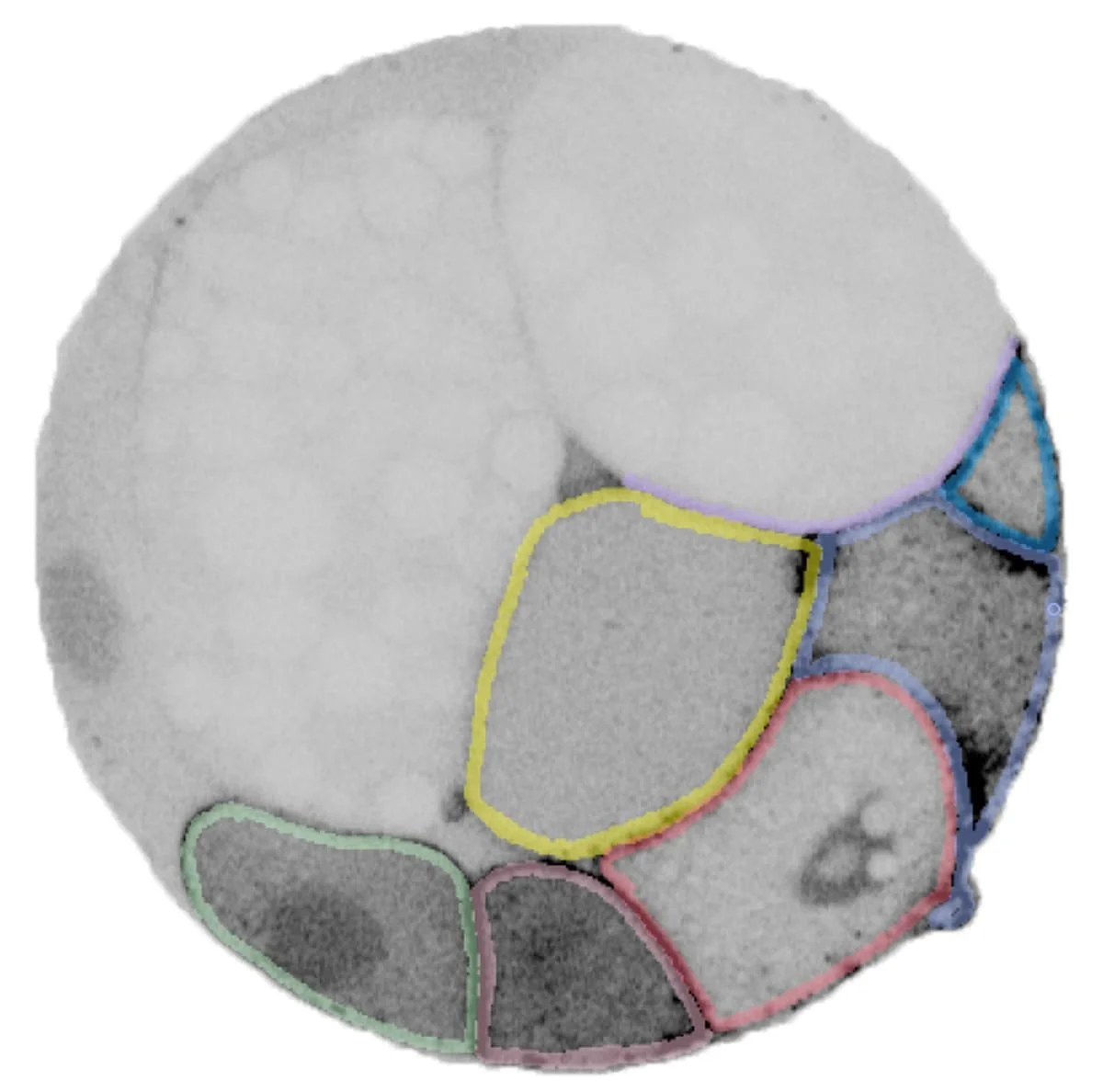
Segmentation of an 32-cell Platynereis dumerilii embryo. The confocal microscope image on the right is one of a z-stack used to create the 3D model of the same embryo above.
I am fascinated by how what we can know about the natural world is inseparable from how we see it. At the microscopic scale, we have no other way to know organisms other than through lens-based instrumentation, significant manipulation of the tissue, and some form of image-making. A confocal time-lapse sequence of a florescent-labeled tissue, an animation of a 3D rendering, or visualization of single-cell RNA sequencing for example, each provides a different view of life at this scale.
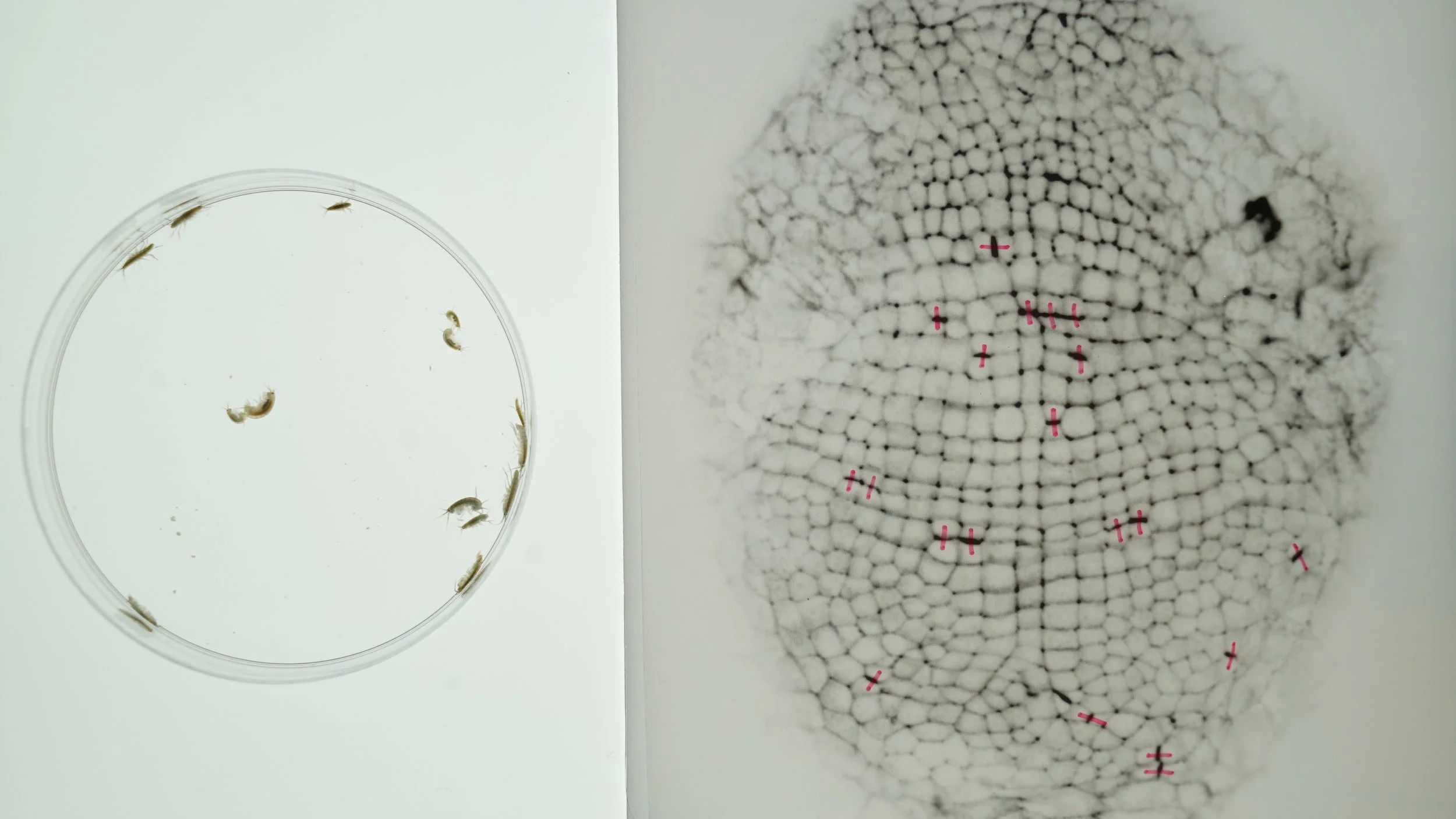
Left: Adult Parhyale hawaiensis. Right: A square grid of cells in the Parhyale hawaiensis embryo (approx. 500 micrometers long). Cells labeled with pink marks are in the process of dividing. See my recent preprint on the morphogenesis of this tissue here.
Much of developmental biology involves grappling with the nuances of microscopic change over time and through cellular space. I love thinking about how to capture, comprehend, and represent the movement of cells or sub-cellular structures with microscopes and various image-analysis modalities. My work is centered around long-term live-imaging with confocal and lightsheet microscopy and mechanical and molecular perturbations to understand morphogenetic mechanisms.
Cellular development in the Parhyale germ band, the embryonic structure that establishes the blueprint for the adult body. On the left is a 16-cell stage embryo and on the right is the same embryo approximately two days later. This time-lapse was take on a multiview lightsheet microscope.
Movement of neuronal mitochondria in a Drosophila melanogaster larva as a time-lapse animation (left) and kymograph (right). I produced this timelapse on a 2-photon microscope to visualize the effect of neurodegenerative mutations on mitochondria morphogenesis in sensory neurons.
I have found that while advanced image analysis, including machine-learning based modalities, now allows us to see these processes in novel and exciting ways, it is also in many ways upending the embodied observational practices that have been central to biology for centuries. To address this, I am exploring ways of using virtual reality to engage with microscope image datasets.
Visualizing Crepidula fornicata embryos in VR and under the microscope
Video produced by Hyacinth Empinado and STAT News (2017)
In 2018, I co-organized the Embryo Journal Club at the Marine Biological Laboratory in Woods Hole, MA. Our aim was to rediscover classical studies on marine invertebrate development and explore new approaches to addressing old questions. We made our process and findings available through our blog and twitter.